library(tidyverse)
library(stringi)
library(scales)
library(gt)Overview
During my (ongoing) job search for a data science or developer-focused role where I get to do R programming, this question came to me: Just how many R developers are there? That’s the question that inspired this post. However, the data needed to answer this question can also be used to answer other interesting questions about R developers, such as how many packages they’ve contributed to, their roles in package development, and so forth. So that’s what we’ll be doing here.
If you just want to see the stats, you can skip to the R developer statistics section. Otherwise follow along to see how I retrieved and wrangled the data into a usable state.
Prerequisites
I’ll be using the CRAN package repository data returned by tools::CRAN_package_db() to get package and author metadata for the current packages available on CRAN. This returns a data frame with character columns containing most metadata from the DESCRIPTION file of a given R package.
Since this data will change over time, here’s when tools::CRAN_package_db() was run for reference: 2023-05-03.
cran_pkg_db <- tools::CRAN_package_db()
glimpse(cran_pkg_db)#> Rows: 19,473
#> Columns: 67
#> $ Package <chr> "A3", "AalenJohansen", "AATtools", "ABACUS",…
#> $ Version <chr> "1.0.0", "1.0", "0.0.2", "1.0.0", "0.1", "0.…
#> $ Priority <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ Depends <chr> "R (>= 2.15.0), xtable, pbapply", NA, "R (>=…
#> $ Imports <chr> NA, NA, "magrittr, dplyr, doParallel, foreac…
#> $ LinkingTo <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, "Rcp…
#> $ Suggests <chr> "randomForest, e1071", "knitr, rmarkdown", N…
#> $ Enhances <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ License <chr> "GPL (>= 2)", "GPL (>= 2)", "GPL-3", "GPL-3"…
#> $ License_is_FOSS <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ License_restricts_use <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ OS_type <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ Archs <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ MD5sum <chr> "027ebdd8affce8f0effaecfcd5f5ade2", "d7eb2a6…
#> $ NeedsCompilation <chr> "no", "no", "no", "no", "no", "no", "no", "n…
#> $ Additional_repositories <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ Author <chr> "Scott Fortmann-Roe", "Martin Bladt [aut, cr…
#> $ `Authors@R` <chr> NA, "c(person(\"Martin\", \"Bladt\", email =…
#> $ Biarch <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ BugReports <chr> NA, NA, "https://github.com/Spiritspeak/AATt…
#> $ BuildKeepEmpty <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ BuildManual <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ BuildResaveData <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ BuildVignettes <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ Built <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ ByteCompile <chr> NA, NA, "true", NA, NA, NA, NA, NA, NA, NA, …
#> $ `Classification/ACM` <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ `Classification/ACM-2012` <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ `Classification/JEL` <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ `Classification/MSC` <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ `Classification/MSC-2010` <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ Collate <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ Collate.unix <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ Collate.windows <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ Contact <chr> NA, NA, NA, NA, NA, NA, NA, NA, "Ian Morison…
#> $ Copyright <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, "Eli…
#> $ Date <chr> "2015-08-15", NA, NA, NA, "2021-12-12", NA, …
#> $ `Date/Publication` <chr> "2015-08-16 23:05:52", "2023-03-01 10:42:09 …
#> $ Description <chr> "Supplies tools for tabulating and analyzing…
#> $ Encoding <chr> NA, "UTF-8", "UTF-8", "UTF-8", "UTF-8", NA, …
#> $ KeepSource <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ Language <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ LazyData <chr> NA, NA, "true", "true", NA, "true", NA, NA, …
#> $ LazyDataCompression <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ LazyLoad <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, "yes", N…
#> $ MailingList <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ Maintainer <chr> "Scott Fortmann-Roe <scottfr@berkeley.edu>",…
#> $ Note <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ Packaged <chr> "2015-08-16 14:17:33 UTC; scott", "2023-02-2…
#> $ RdMacros <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ StagedInstall <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ SysDataCompression <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ SystemRequirements <chr> NA, NA, NA, NA, NA, NA, NA, NA, "GNU make", …
#> $ Title <chr> "Accurate, Adaptable, and Accessible Error M…
#> $ Type <chr> "Package", "Package", "Package", NA, "Packag…
#> $ URL <chr> NA, NA, NA, "https://shiny.abdn.ac.uk/Stats/…
#> $ UseLTO <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ VignetteBuilder <chr> NA, "knitr", NA, "knitr", NA, "knitr", NA, N…
#> $ ZipData <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ Path <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ `X-CRAN-Comment` <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ Published <chr> "2015-08-16", "2023-03-01", "2022-08-12", "2…
#> $ `Reverse depends` <chr> NA, NA, NA, NA, NA, NA, "abctools, EasyABC",…
#> $ `Reverse imports` <chr> NA, NA, NA, NA, NA, NA, "ecolottery, poems",…
#> $ `Reverse linking to` <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ `Reverse suggests` <chr> NA, NA, NA, NA, NA, NA, "coala", "abctools",…
#> $ `Reverse enhances` <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …Wrangle
Since we only care about package and author metadata, a good first step is to remove everything else. This leaves us with a Package field and two author fields: Author and Authors@R. The difference between the two author fields is that Author is an unstructured text field that can contain any text in any format, and Authors@R is a structured text field containing R code that defines authors’ names and roles with the person() function.
cran_pkg_db <- cran_pkg_db |>
select(package = Package, authors = Author, authors_r = `Authors@R`) |>
as_tibble()Here’s a comparison of the two fields, using the dplyr package as an example:
# Author
cran_pkg_db |>
filter(package == "dplyr") |>
pull(authors) |>
cat()#> Hadley Wickham [aut, cre] (<https://orcid.org/0000-0003-4757-117X>),
#> Romain François [aut] (<https://orcid.org/0000-0002-2444-4226>),
#> Lionel Henry [aut],
#> Kirill Müller [aut] (<https://orcid.org/0000-0002-1416-3412>),
#> Davis Vaughan [aut] (<https://orcid.org/0000-0003-4777-038X>),
#> Posit Software, PBC [cph, fnd]# Authors@R
cran_pkg_db |>
filter(package == "dplyr") |>
pull(authors_r) |>
cat()#> c(
#> person("Hadley", "Wickham", , "hadley@posit.co", role = c("aut", "cre"),
#> comment = c(ORCID = "0000-0003-4757-117X")),
#> person("Romain", "François", role = "aut",
#> comment = c(ORCID = "0000-0002-2444-4226")),
#> person("Lionel", "Henry", role = "aut"),
#> person("Kirill", "Müller", role = "aut",
#> comment = c(ORCID = "0000-0002-1416-3412")),
#> person("Davis", "Vaughan", , "davis@posit.co", role = "aut",
#> comment = c(ORCID = "0000-0003-4777-038X")),
#> person("Posit Software, PBC", role = c("cph", "fnd"))
#> )And a glimpse at the data:
cran_pkg_db#> # A tibble: 19,473 × 3
#> package authors autho…¹
#> <chr> <chr> <chr>
#> 1 A3 "Scott Fortmann-Roe" <NA>
#> 2 AalenJohansen "Martin Bladt [aut, cre],\n Christian Furrer [aut]" "c(per…
#> 3 AATtools "Sercan Kahveci [aut, cre]" "perso…
#> 4 ABACUS "Mintu Nath [aut, cre]" <NA>
#> 5 abbreviate "Sigbert Klinke [aut, cre]" "\n p…
#> 6 abbyyR "Gaurav Sood [aut, cre]" "perso…
#> 7 abc "Csillery Katalin [aut],\n Lemaire Louisiane [aut],\n… "c( \n…
#> 8 abc.data "Csillery Katalin [aut],\n Lemaire Louisiane [aut],\n… "c( \n…
#> 9 ABC.RAP "Abdulmonem Alsaleh [cre, aut], Robert Weeks [aut], Ia… <NA>
#> 10 ABCanalysis "Michael Thrun, Jorn Lotsch, Alfred Ultsch" <NA>
#> # … with 19,463 more rows, and abbreviated variable name ¹authors_rFrom the output above you can see that every package uses the Author field, but not all packages use the Authors@R field. This is unfortunate, because it means that the names and roles of authors need to be extracted from the unstructured text in the Author field for a subset of packages, which is difficult to do and somewhat error-prone. Just for consideration, here’s how many packages don’t use the Authors@R field.
cran_pkg_db |>
filter(is.na(authors_r)) |>
nrow()#> [1] 6361So roughly one-third of all packages. From the output above it’s also clear that although there are similarities in how different packages populate the Author field, it does vary; so a simple rule like splitting the text on commas isn’t sufficient. These are fairly tame examples—some packages even use multiple sentences describing each author’s roles and affiliations, or contain other comments such as copyright disclaimers. All of these things make it more difficult to extract names and roles without errors.
Conversely, for the Authors@R field, all that’s needed is to parse and evaluate the R code stored there as a character string; this will return a person vector that has format() methods to get authors’ names and roles into an analysis-ready format. This removes the possibility for me to introduce errors into the data, although it doesn’t solve things like Authors using an inconsistent name across packages (e.g., sometimes including their middle initial and sometimes not, or just generally writing their name differently).
Because there are two fields, I’ll make two helper functions to get name and role data from each field. Regardless of the field, the end goal is to tidy cran_pkg_db into a data frame with three columns: package, person, and roles, with one package/person combination per row.
Extracting roles
From the example dplyr output above, we can see that the roles column is currently a character string with the role codes, which isn’t super useful. Later on I’ll split these out into indicator columns with a TRUE or FALSE for whether someone had a given role. I also wanted the full names for the roles, since some of the codes aren’t very obvious.
Kurt Hornik, Duncan Murdoch and Achim Zeileis published a nice article in The R Journal explaining the roles of R package authors and where they come from. Briefly, they come from the “Relator and Role” codes and terms from MARC (MAchine-Readable Cataloging, Library of Congress, 2012) here: https://www.loc.gov/marc/relators/relaterm.html.
There are a lot of roles there; I just took the ones that were present in the data at the time I wrote this post.
marc_roles <- c(
analyst = "anl",
architecht = "arc",
artist = "art",
author = "aut",
author_in_quotations = "aqt",
author_of_intro = "aui",
bibliographic_antecedent = "ant",
collector = "col",
compiler = "com",
conceptor = "ccp",
conservator = "con",
consultant = "csl",
consultant_to_project = "csp",
contestant_appellant = "cot",
contractor = "ctr",
contributor = "ctb",
copyright_holder = "cph",
corrector = "crr",
creator = "cre",
data_contributor = "dtc",
degree_supervisor = "dgs",
editor = "edt",
funder = "fnd",
illustrator = "ill",
inventor = "inv",
lab_director = "ldr",
lead = "led",
metadata_contact = "mdc",
musician = "mus",
owner = "own",
presenter = "pre",
programmer = "prg",
project_director = "pdr",
scientific_advisor = "sad",
second_party = "spy",
sponsor = "spn",
supporting_host = "sht",
teacher = "tch",
thesis_advisor = "ths",
translator = "trl",
research_team_head = "rth",
research_team_member = "rtm",
researcher = "res",
reviewer = "rev",
witness = "wit",
woodcutter = "wdc"
)Tidying the data
With all the explanations out of the way we can now tidy the data with our helper functions.
cran_authors <- cran_pkg_db |>
mutate(
# Letters with accents, etc. should be normalized so that names including
# them are picked up by the regex.
across(c(authors, authors_r), \(.x) stri_trans_general(.x, "latin-ascii")),
# The extraction functions aren't vectorized so they have to be mapped over.
# This creates a list column.
persons = if_else(
is.na(authors_r),
map(authors, \(.x) authors(.x)),
map(authors_r, \(.x) authors_r(.x))
)
) |>
select(-c(authors, authors_r)) |>
unnest(persons) |>
# If a package only has one author then they must be the author and creator,
# so it's safe to impute this when it isn't there.
group_by(package) |>
mutate(roles = if_else(
is.na(roles) & n() == 1, "[aut, cre]", roles
)) |>
ungroup()Then add the indicator columns for roles. Note the use of the walrus operator (:=) here to create new columns from the full names of MARC roles on the left side of the walrus, while detecting the MARC codes with str_detect() on the right side. I’m mapping over this because the left side can’t be a vector.
cran_authors_tidy <- cran_authors |>
# Add indicator columns for all roles.
bind_cols(
map2_dfc(
names(marc_roles), marc_roles,
function(.x, .y) {
cran_authors |>
mutate(!!.x := str_detect(roles, .y)) |>
select(!!.x)
}
)
) |>
# Not everyone's role is known.
mutate(unknown = is.na(roles))This all leaves us with a tidy (mostly error free) data frame about R developers and their roles that is ready to explore:
glimpse(cran_authors_tidy)#> Rows: 52,719
#> Columns: 50
#> $ package <chr> "A3", "AalenJohansen", "AalenJohansen", "AATt…
#> $ person <chr> "Scott Fortmann-Roe", "Martin Bladt", "Christ…
#> $ roles <chr> "[aut, cre]", "[aut, cre]", "[aut]", "[aut, c…
#> $ analyst <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ architecht <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ artist <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ author <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRU…
#> $ author_in_quotations <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ author_of_intro <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ bibliographic_antecedent <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ collector <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ compiler <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ conceptor <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ conservator <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ consultant <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ consultant_to_project <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ contestant_appellant <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ contractor <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ contributor <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ copyright_holder <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ corrector <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ creator <lgl> TRUE, TRUE, FALSE, TRUE, TRUE, TRUE, TRUE, FA…
#> $ data_contributor <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ degree_supervisor <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ editor <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ funder <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ illustrator <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ inventor <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ lab_director <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ lead <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ metadata_contact <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ musician <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ owner <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ presenter <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ programmer <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ project_director <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ scientific_advisor <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ second_party <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ sponsor <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ supporting_host <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ teacher <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ thesis_advisor <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ translator <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ research_team_head <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ research_team_member <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ researcher <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ reviewer <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ witness <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ woodcutter <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
#> $ unknown <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FAL…R developer statistics
I’ll start with person-level stats, mainly because some of the other stats are further summaries of these statistics. Nothing fancy here, just the number of packages a person has contributed to, role counts, and nominal and percentile rankings. Both the ranking methods used here give every tie the same (smallest) value, so if two people tied for second place both their ranks would be 2, and the next person’s rank would be 4.
cran_author_pkg_counts <- cran_authors_tidy |>
group_by(person) |>
summarise(
n_packages = n(),
across(analyst:unknown, function(.x) sum(.x, na.rm = TRUE))
) |>
mutate(
# Discretizing this for visualization purposes later on
n_pkgs_fct = case_when(
n_packages == 1 ~ "One",
n_packages == 2 ~ "Two",
n_packages == 3 ~ "Three",
n_packages >= 4 ~ "Four+"
),
n_pkgs_fct = factor(n_pkgs_fct, levels = c("One", "Two", "Three", "Four+")),
rank = min_rank(desc(n_packages)),
percentile = percent_rank(n_packages) * 100,
.after = n_packages
) |>
arrange(desc(n_packages))Here’s an interactive gt table of the person-level stats so you can find yourself, or ask silly questions like how many other authors share a name with you. If you page or search through it you can also get an idea of the data quality (e.g., try “Posit” under the person column and you’ll see that they don’t use a consistent organization name across all packages, which creates some measurement error here).
Code
cran_author_pkg_counts |>
select(-n_pkgs_fct) |>
gt() |>
tab_header(
title = "R Developer Contributions",
subtitle = "CRAN Package Authorships and Roles"
) |>
text_transform(
\(.x) str_to_title(str_replace_all(.x, "_", " ")),
locations = cells_column_labels()
) |>
fmt_number(
columns = percentile
) |>
fmt(
columns = rank,
fns = \(.x) label_ordinal()(.x)
) |>
cols_width(everything() ~ px(120)) |>
opt_interactive(use_sorting = FALSE, use_filters = TRUE)So there are around 29453 people who have some type of authorship on at least one currently available CRAN package at the time this post was published. I’ve emphasized “around” because of the measurement error from extracting names from the Author field of DESCRIPTION and from people writing their names in multiple ways across packages, but also because this number will fluctuate over time as new packages are published, unmaintained packages are archived, and so forth.
To try to put this number into perspective, Ben Ubah, Claudia Vitolo, and Rick Pack put together a dashboard with data on how many R users there are worldwide belonging to different R user groups. At the time of writing this post there were:
- Around 775,000 members of R user groups organized on Meetup
- Around 100,000 R-Ladies members
The R Consortium also states on their website that there are more than two million R users worldwide (although they don’t state when or where this number comes from). Regardless of the exact amount, it’s apparent that there are many more R users than R developers.
Package contributions
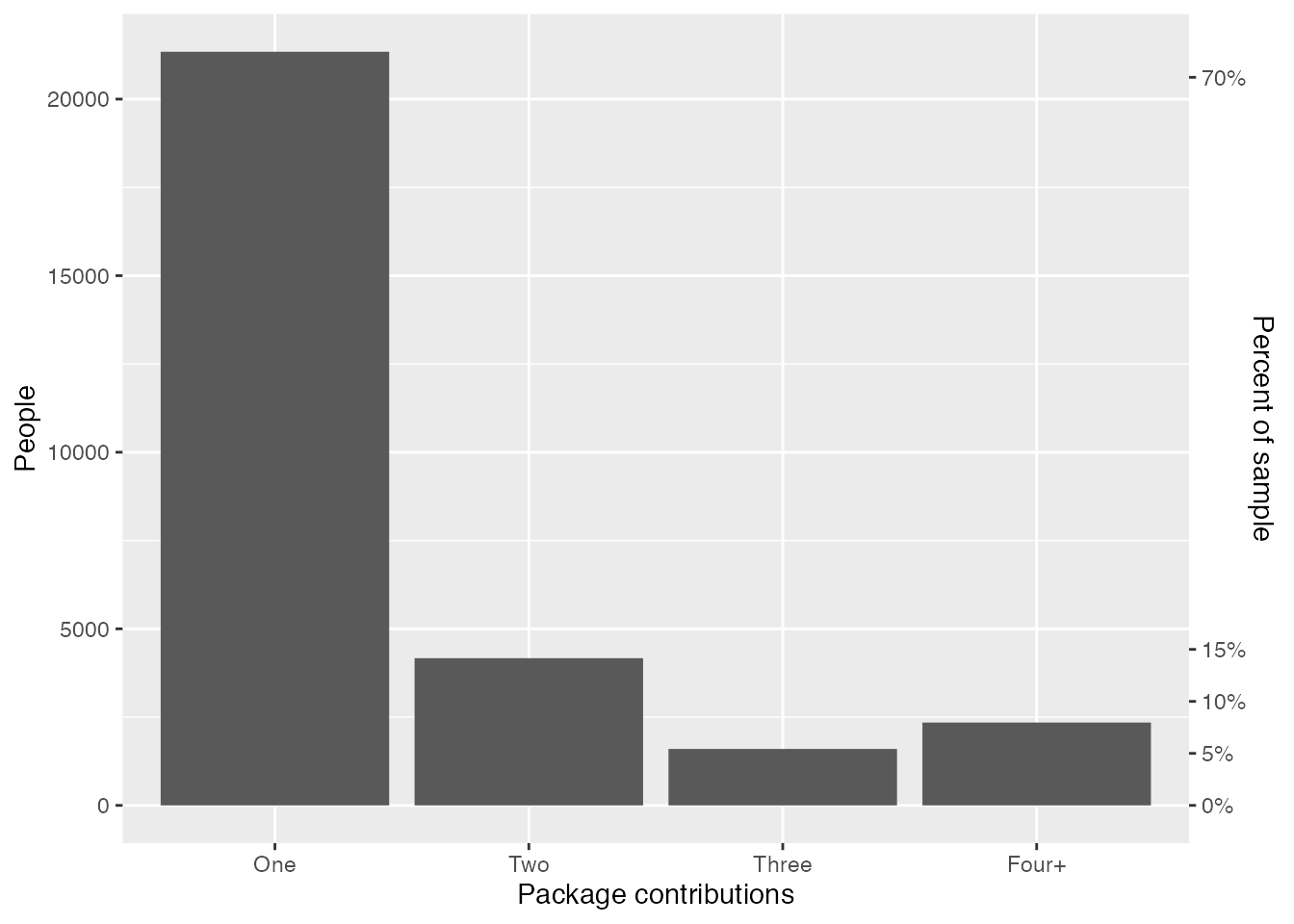
The title of this post probably gave this away, but around 90% of R developers have worked on one to three packages, and only around 10% have worked on four or more packages.
cran_author_pkg_counts |>
group_by(n_pkgs_fct) |>
summarise(n_people = n()) |>
ggplot(mapping = aes(x = n_pkgs_fct, y = n_people)) +
geom_col() +
scale_y_continuous(
sec.axis = sec_axis(
trans = \(.x) .x / nrow(cran_author_pkg_counts),
name = "Percent of sample",
labels = label_percent(),
breaks = c(0, .05, .10, .15, .70)
)
) +
labs(
x = "Package contributions",
y = "People"
)
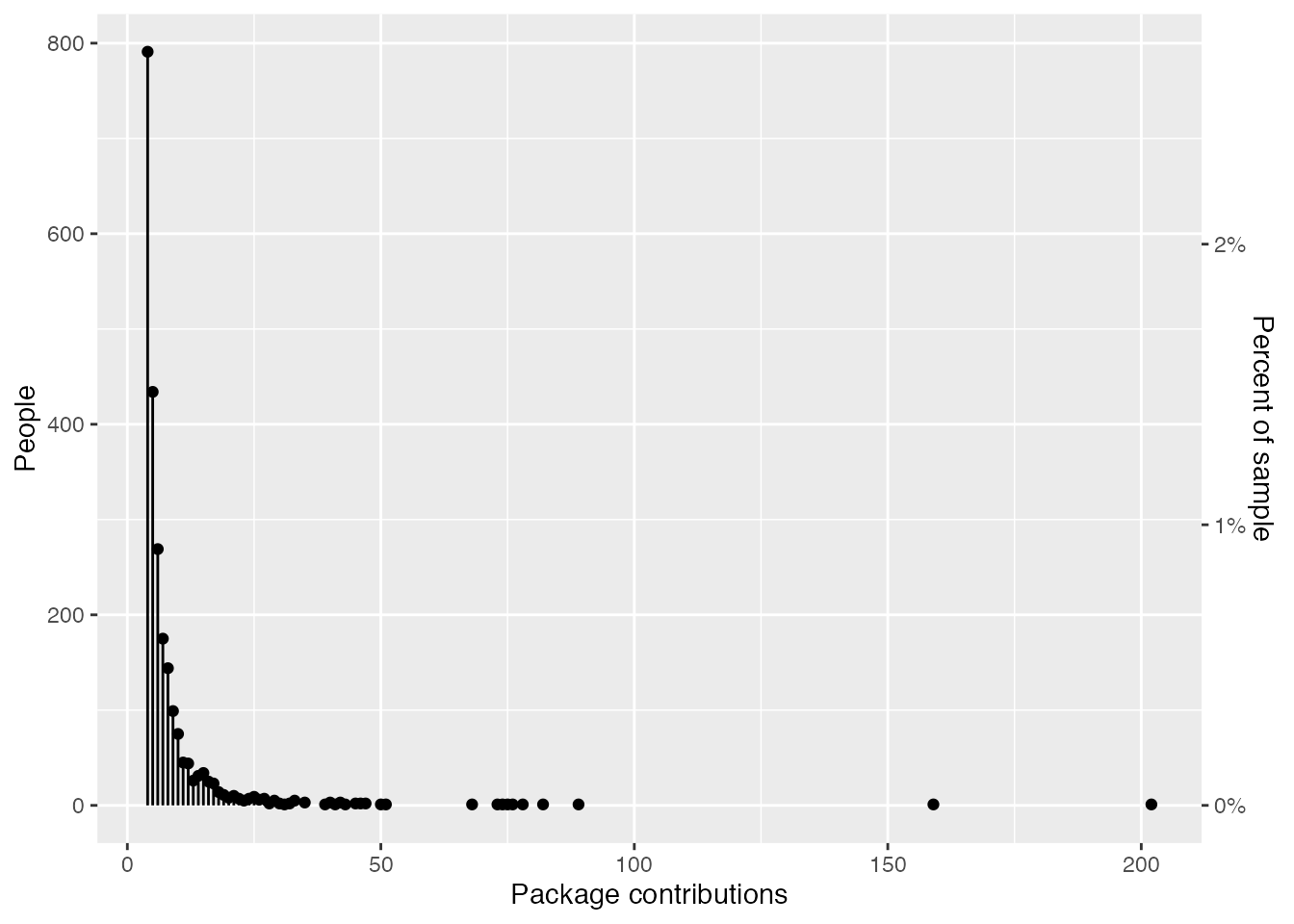
Notably, in the group that have worked on four or more packages, the spread of package contributions is huge. This vast range is mostly driven by people who do R package development as part of their job (e.g., if you look at the cran_author_pkg_counts table above, most of the people at the very top are either professors of statistics or current or former developers from Posit, rOpenSci, or the R Core Team).
cran_author_pkg_counts |>
filter(n_pkgs_fct == "Four+") |>
group_by(rank, n_packages) |>
summarise(n_people = n()) |>
ggplot(mapping = aes(x = n_packages, y = n_people)) +
geom_segment(aes(xend = n_packages, yend = 0)) +
geom_point() +
scale_y_continuous(
sec.axis = sec_axis(
trans = \(.x) .x / nrow(cran_author_pkg_counts),
name = "Percent of sample",
labels = label_percent()
)
) +
labs(
x = "Package contributions",
y = "People"
)
Here are some subsample summary statistics to compliment the plots above.
cran_author_pkg_counts |>
group_by(n_packages >= 4) |>
summarise(
n_developers = n(),
n_pkgs_mean = mean(n_packages),
n_pkgs_sd = sd(n_packages),
n_pkgs_median = median(n_packages),
n_pkgs_min = min(n_packages),
n_pkgs_max = max(n_packages)
)#> # A tibble: 2 × 7
#> `n_packages >= 4` n_developers n_pkgs_mean n_pkgs_sd n_pkgs_…¹ n_pkg…² n_pkg…³
#> <lgl> <int> <dbl> <dbl> <dbl> <int> <int>
#> 1 FALSE 27107 1.27 0.562 1 1 3
#> 2 TRUE 2346 7.78 8.63 5 4 202
#> # … with abbreviated variable names ¹n_pkgs_median, ²n_pkgs_min, ³n_pkgs_maxRole distributions
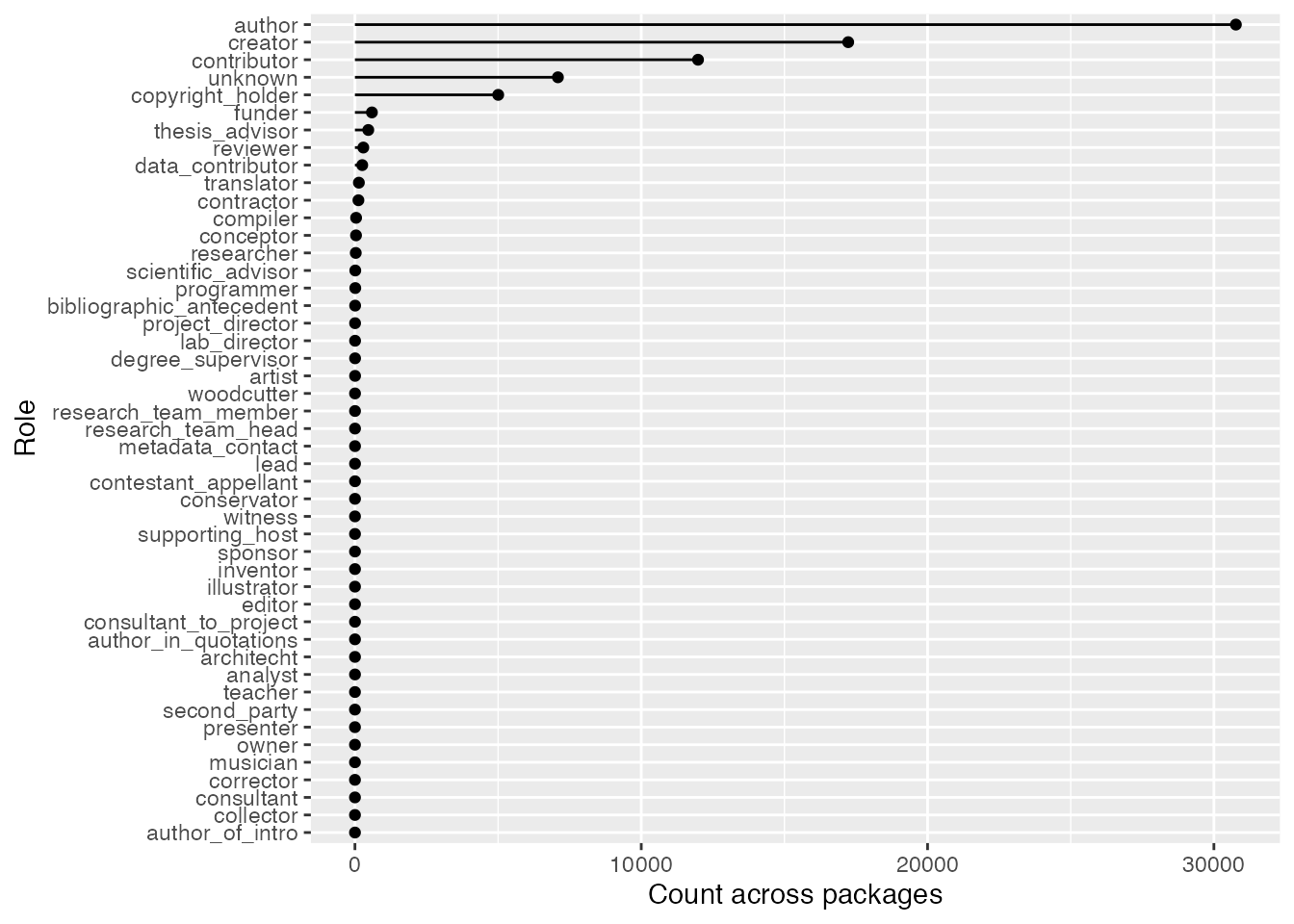
Not every contribution to an R package involves code. For example, two authors of the wiad package were woodcutters! The package is for wood image analysis, so although it’s surprising a role like that exists, it makes a lot of sense in context. Anyways, neat factoids aside, the point of this section is to look at the distribution of different roles in R package development.
To start, let’s get an idea of how many people were involved in programming-related roles. This won’t be universally true, but most of the time the following roles will involve programming:
programming_roles <-
c("author", "creator", "contributor", "compiler", "programmer")Here’s the count:
cran_author_pkg_counts |>
filter(if_any(!!programming_roles, \(.x) .x > 0)) |>
nrow()#> [1] 24170There were also 5434 whose role was unknown (either because it wasn’t specified or wasn’t picked up by my regex method). Regardless, most people have been involved in programming-related roles, and although other roles occur they’re relatively rare.
Here’s a plot to compliment this point:
cran_authors_tidy |>
summarise(across(analyst:unknown, function(.x) sum(.x, na.rm = TRUE))) |>
pivot_longer(cols = everything(), names_to = "role", values_to = "n") |>
arrange(desc(n)) |>
ggplot(mapping = aes(x = n, y = reorder(role, n))) +
geom_segment(aes(xend = 0, yend = role)) +
geom_point() +
labs(
x = "Count across packages",
y = "Role"
)
Ranking contributions
The interactive table above already contains this information, but to compliment David Smith’s post from 5 years ago, here’s the current Top 20 most prolific authors on CRAN.
This is why Hadley is on the cover of Glamour magazine and we’re not.

cran_author_pkg_counts |>
# We don't want organizations or groups here
filter(!(person %in% c("RStudio", "R Core Team", "Posit Software, PBC"))) |>
head(20) |>
select(person, n_packages) |>
gt() |>
tab_header(
title = "Top 20 R Developers",
subtitle = "Based on number of CRAN package authorships"
) |>
text_transform(
\(.x) str_to_title(str_replace_all(.x, "_", " ")),
locations = cells_column_labels()
) |>
cols_width(person ~ px(140))| Top 20 R Developers | |
| Based on number of CRAN package authorships | |
| Person | N Packages |
|---|---|
| Hadley Wickham | 159 |
| Jeroen Ooms | 89 |
| Gabor Csardi | 82 |
| Kurt Hornik | 78 |
| Scott Chamberlain | 76 |
| Dirk Eddelbuettel | 75 |
| Martin Maechler | 74 |
| Stephane Laurent | 73 |
| Achim Zeileis | 68 |
| Winston Chang | 51 |
| Max Kuhn | 50 |
| Yihui Xie | 47 |
| Jim Hester | 46 |
| Henrik Bengtsson | 45 |
| John Muschelli | 45 |
| Roger Bivand | 43 |
| Ben Bolker | 42 |
| Bob Rudis | 42 |
| Brian Ripley | 42 |
| Michel Lang | 41 |
Conclusion
My main takeaway from all of this is that if you know how to write and publish an R package on CRAN (or contribute to existing packages), you have a valuable skill that not a lot of other R users have. If you do want to learn, I recommend reading R Packages by Hadley Wickham and Jenny Bryan.
My other takeaway is that the Author field should be dropped from DESCRIPTION so my eyesore of a regular expression never has to extract a name again. (This still wouldn’t remove all the measurement error I discussed, since some people and organizations don’t write their names consistently across packages. Oh well.).
One thing I am curious about, but which would be hard to get good data on, is how many people have R package development experience who haven’t published on CRAN; or, of the people who have published on CRAN, how many packages have they worked on that aren’t (yet) on CRAN (for me it’s five).
Anyways, that’s it for now. If you think this data could answer other interesting questions I didn’t cover, let me know down below and I’ll consider adding more to the post.

Michael McCarthy
Thanks for reading! I’m Michael, the voice behind Tidy Tales. I am an award winning data scientist and R programmer with the skills and experience to help you solve the problems you care about. You can learn more about me, my consulting services, and my other projects on my personal website.
Session Info
─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.2.2 (2022-10-31)
os macOS Mojave 10.14.6
system x86_64, darwin17.0
ui X11
language (EN)
collate en_CA.UTF-8
ctype en_CA.UTF-8
tz America/Vancouver
date 2023-05-03
pandoc 2.14.0.3 @ /Applications/RStudio.app/Contents/MacOS/pandoc/ (via rmarkdown)
quarto 1.2.313 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
dplyr * 1.1.0 2023-01-29 [1] CRAN (R 4.2.0)
forcats * 0.5.2 2022-08-19 [1] CRAN (R 4.2.0)
ggplot2 * 3.4.0 2022-11-04 [1] CRAN (R 4.2.0)
gt * 0.9.0 2023-03-31 [1] CRAN (R 4.2.0)
purrr * 0.3.5 2022-10-06 [1] CRAN (R 4.2.0)
readr * 2.1.3 2022-10-01 [1] CRAN (R 4.2.0)
scales * 1.2.1 2022-08-20 [1] CRAN (R 4.2.0)
sessioninfo * 1.2.2 2021-12-06 [1] CRAN (R 4.2.0)
stringi * 1.7.8 2022-07-11 [1] CRAN (R 4.2.0)
stringr * 1.5.0 2022-12-02 [1] CRAN (R 4.2.0)
tibble * 3.1.8 2022-07-22 [1] CRAN (R 4.2.0)
tidyr * 1.2.1 2022-09-08 [1] CRAN (R 4.2.0)
tidyverse * 1.3.2 2022-07-18 [1] CRAN (R 4.2.0)
[1] /Users/Michael/Library/R/x86_64/4.2/library/__tidytales
[2] /Library/Frameworks/R.framework/Versions/4.2/Resources/library
──────────────────────────────────────────────────────────────────────────────Citation
@online{mccarthy2023,
author = {Michael McCarthy},
title = {The {Pareto} {Principle} in {R} Package Development},
date = {2023-05-03},
url = {https://tidytales.ca/posts/2023-05-03_r-developers},
langid = {en}
}
Comments